Halloski!
Welcome to my personal website - I am Magdalena and I work as a Postdoctoral Fellow in the group of Prof. Mark Tester in KAUST, Saudi Arabia. I am a plant physiologist interested in how plant architecture affects plant tolerance to different abiotic stresses. I pick my favourite genes, affecting plant architecture, based on the forward genetic studies (GWAS), as well as RNAseq results from lines differing in salinity tolerance and root architecture. I develop simple models describing the change in the individual elements of plant architecture throughout time and use them to simplify the complexity of the entire plant. I like to share the developed pipelines for data analysis in the form of the shiny apps and data analysis pipelines (see CURRENT PROJECTS below).
MY BIO:
I was born in Zyrardow, a small industrial city in Poland build with red-bricks. My parents moved to the Netherlands, where I finished my high-school and started my intellectual germination at the University of Amsterdam. There I got my BSc degree in General Biology and MSc degree in Green Life Science. During my MSc degree I did two fun MSc projects, one in Amsterdam, focusing on SnRK2.4/2.10 interaction with phospholipids and the other one in Canberra, on blue-light induced processes leading to germination of wheat.
Later, my seedling brain was nurtured by Prof. Christa Testerink, during my PhD project at University of Amsterdam. I studied Root System Architecture of Arabidopsis HapMap population, developed a simple Root-Fit model to describe stress-induced reprogramming of the architecture, and explored the salt stress induced changes in the rosette size and identified a new gene LRR-KISS to be important for growth maintenance under saline conditions. During my PhD I also studied on protein-lipid interactions using liposome binding assays focusing on the interactions between Phosphatidic Acid and Arabidopsis SnRK2.4/2.10 proteins. Together with (back then) BSc. student Elmer Swart we collected some Arabidopsis accessions throughout the Netherlands, to examine their salinity tolerance.

Right now I am having a post-doc fun in KAUST Saudi Arabia. I am focusing on (1) salt-induced changes in root-to-shoot ratio in Arabidopsis, (2) study the expression patterns in plants with enhanced sodium accumulation in their roots and (3) developing tools for data analysis based on R/Shiny. I am passionate about capturing plant architecture using simple models, understanding plant physiology and salinity tolerance. I love coding in R, BIG data analysis and sharing whatever I know with whomever cares to listen. I am also quite proficient in classic molecular biology and high-throughput phenotyping. Occasionally, I join for fieldwork - either looking for native Saudi Arabidopsis, or digging out the tomatoes from the field trials to look at their roots.

I am an active reviewer for plant science journals, a reviewing editor at Frontiers in Plant Science and a Plantae Fellow writting small paper summaries for “What we’re reading” series, which appears weakly on the Plantae website. You can find the bits that I write here.

Working at KAUST gives me a unique opportunity to experience culture and landscape that is very different from my own. You can read about my experiences of the Middle East on my blog.

MY RESEARCH:
Plants are possibly the best system to study the interactions between an organism and its environment. As a student, I was captured by the ability of plants to sense changing environment and translate it to molecular signals resulting in altered cell division patterns, differentiation of the meristem, which ultimately lead to a different plant architecture that can be studied with a naked eye. Investment in new tissues is initially energetically expensive, but increases plant productivity by adding new source tissues, such as leaves or roots.
My hypothesis is that plants able to adapt their architecture most effectively to changing environments at individual life stages show enhanced survival and productivity, resulting in evolutionary advantage. The integration of abiotic stress signals is especially important, as the resources become limiting.

Salt stress induces quiescence of growth through modification of cell cycle activity, cell expansion and cell wall extensibility. The period of initial growth arrest and the extent of recovered growth differs between individual organs leading to altered plant morphology. So far we studied altered morphology of Root System Architecture, by capturing growth dynamics in simple descriptive models, like Root-Fit. By examining natural variation therein we identified four distinct responses, which correspond to maintenance of ion equilibrium in shoot tissue. When Root System Architecture data was used for Genome Wide Association Studies, we were able to identify a number of novel candidate genes as well as genes previously linked to salinity tolerance, for which we unravelled their role in root development. Our results show how study of growth dynamics lead to novel plant trait discovery, which is especially relevant in these times of fast development in plant phenotyping approaches.
CURRENT PROJECTS:
## MVApp

Plants are master shape-shifters, constantly adapting their architecture to enhance resource acquisition, ensuring reproductive success. These metamorphoses are governed by genotype and environment (GxE) interactions. Understanding the full range of plant’s morphological versatility, environmental triggers and adaptive relevance often requires a thorough assessment of large numbers of genotypes under various conditions. Recent advances in phenotyping technologies allow the high-throughput quantification of traits reflecting plant size, shape, photosynthetic efficiency and transpiration rate. With increasing number of phenotypes it is becoming increasingly crucial, and proportionally complicated, to identify redundancy, correlations, and impact in phenotypic datasets.
To adress all of the above, together with a group of five PhD students at KAUST, we developed the MVApp, a user-friendly multivariate analysis tool for large or small datasets.
The MVApp features include:
- Curve fitting for plant growth estimation
- Data curation for possible outlier values
- Data examination for significant differences between genotypes or treatments
- Correlation analysis across different traits
- Principle Component Analysis and Multidimensional Scaling
- Clustering analysis (Hierarchical and K-means)
- Broad sense heritability estimation
- Quantile regression analysis
We hope that the MVApp will be applicable beyond plant science field, contributing to transparent data curation, analysis, and processing, but also serve as a teaching tool for statistical methods for the ones not (yet) familiar with R and Multivariate analysis.
You can find instructions on how to use it here, and if you dont feel like reading we made some YouTube tutorials available here.
## Salt induced changes in root-to-shoot ratio

Notably, salt stress rapidly impacts the growth and development of different plant organs to different extents, leading to alterations in plant development. In Arabidopsis Col-0 genotype, reduction in main root growth is more severe than reduction in lateral root growth or development, resulting in altered root architecture. Other alterations to plant development, such as root-to-shoot ratio are likely to be also contributing to salinity tolerance. In this study, we describe salt stress and hormone induced changes in root-to-shoot ratio in Arabidopsis plants grown on agar plates, by quantifying the dynamics of root and shoot growth in different conditions. We developed a tool that enables estimation of increases in root and shoot mass through quantification of green and white pixels and automated fitting of exponential growth curves. We identified genetic components underlying salt stress-induced changes in root:shoot ratio by studying natural variation in the Arabidopsis “HapMap” population and performing association genetic analyses to identify candidate genes. Further validation of candidate genes will provide more insight into signals regulating stress-induced alterations in plant development and their role in overall salinity tolerance.
## Transcriptional networks controling lateral root development under salinity stress

Previous studies describe that high expression of Arabidopsis High Affinity K+ Transporter (HKT1;1), sequestering sodium from the transpiration stream in root pericycle, results in increased salinity tolerance. Our results show that pericycle specific overexpression of HKT1 results in reduced lateral root development under saline conditions. Interestingly, this effect seems to be specific to dicot species, as wheat lines with high HKT1 expression do not show reduction in secondary lateral roots. In this project we identify the transcriptional regulation of lateral root development downstream of HKT1 through comparative transcriptomic analysis of two lines with stelar overexpression of HKT1. Among the transcripts that were expressed differentially between HKT1 overexpression lines and their background lines, we found genes involved in cell differentiation, ABA signaling and transport. Further validation of identified candidate genes will enhance our understanding of processes controlling plant development under abiotic stress conditions and the importance of root architecture for salinity tolerance.
## SNPer - precision GWAS mapping

The Forward genetic screens are a great tool to establish the links between the genetic code and the observed phenotype. However, this can get a little messy, when you are using different models and many different traits as input for your mapping. As the number of SNPs that you can use for mapping is increasing, it is often very difficult to make a visual inspection of the mapping. Using SNPer, you can inspect the GWAS results for multiple traits at the same time and examine the overlapping associations between different traits at the resolution that you want - due to the interactive plotly graphs. The tool is available here, instructions on how to use it are here, while the code for the SNPer is available here. DOI: 10.5281/zenodo.1227775
## Salt induced changes in RSA

While everyone knows that salt stress reduces root growth, salt stress induced changes in Root System Architecture are less studied. During my PhD, I examined natural variation in salt stress by studying the root architectures of 347 Arabidopsis accessions. It was a lot of work! We collected 17 RSA traits in three different conditions, and used this data for Genome Wide Association Study (GWAS). Our GWAS identified 100 loci, among which CYP79B2 and HKT1. CYP79B2 was validated to be involved in maintenance of lateral root growth during salt stress, possibly by altering auxin biosynthesis. Enhanced HKT1 expression in root stelle repressed lateral root formation under salt stress conditions, revealing that while retention of salt ions in the root is an excellent mechanism of salinity tolerance in larger plants, it is detrimental for primordia development in young seedlings. The validation of other identified loci remains to be revealed.
The data was integrated into an on-line app, where you can have fun comparing the collected phenotypes of individual accessions, correlating different traits and even doing some clustering analysis based on YOUR favourite traits. The app is available here or here, while the instructions on how to use it are available here.
The paper was published as Julkowska et al., The Plant Cell 2017. DOI: https://doi.org/10.1105/tpc.16.00680
## Natural variation in rosette size

Natural variation among Arabidopsis accessions is broadly recognized as an important genetic resource for identification of mechanisms underlying plant development and stress tolerance. To evaluate the natural variation insalinity stress tolerance, two large-scale experiments were performed on two populations consisting of 160 Arabidopsis accessions each. Traits such as projected rosette area, fresh and dry weight were collected as an estimate for salinity tolerance. The results revealed considerable natural variation in salt stress responses among the accessions studied. The rosette size at salt stress was observed to correspond to the developmental differences observed between the accessions studied at control conditions. Subsequent Genome Wide Association Study (GWAS) revealed associations with novel candidate genes for salinity tolerance such as LRR-KISS, flowering locus KH-domain containing protein and DUF1639 containing protein. The allelic variation underlying LRR-KISS expression is hypothesized to contribute to salinity tolerance by maintaining the rosette growth.
This project was a part of the Ecogenomics course for 3rd year Biology students at University of Amsterdam. The data collection was a real group effort, and I will always be very very thankful for the input and the enthousiasm of those amazing students that I had honour and pleasure to supervise!
The paper was published as Julkowska et al., The Journal of Experimental Botany 2016, 1060, 67 (8), 2127-2138
## Root-Fit

The plant root is the first organ to encounter salinity stress, but the effect of salinity on root system architecture (RSA) remains elusive. Both the reduction in main root (MR) elongation and the redistribution of the root mass between MRs and lateral roots (LRs) are likely to play crucial roles in water extraction efficiency and ion exclusion. To establish which RSA parameters are responsive to salt stress, we performed a detailed time course experiment in which Arabidopsis (Arabidopsis thaliana) seedlings were grown on agar plates under different salt stress conditions. We captured RSA dynamics with quadratic growth functions (root-fit) and summarized the salt-induced differences in RSA dynamics in three growth parameters: MR elongation, average LR elongation, and increase in number of LRs. In the ecotype Columbia-0 accession of Arabidopsis, salt stress affected MR elongation more severely than LR elongation and an increase in LRs, leading to a significantly altered RSA. By quantifying RSA dynamics of 31 different Arabidopsis accessions in control and mild salt stress conditions, different strategies for regulation of MR and LR meristems and root branching were revealed. Different RSA strategies partially correlated with natural variation in abscisic acid sensitivity and different Na(+)/K(+) ratios in shoots of seedlings grown under mild salt stress. Applying root-fit to describe the dynamics of RSA allowed us to uncover the natural diversity in root morphology and cluster it into four response types that otherwise would have been overlooked.
After the publication of the paper, I was approached by Guillaume Lobet, who constructed a shiny-tool for Root-Fit. You can find the GitHub repository here.
The paper was published as Julkowska et al., Plant Physiology 2014, 166:1387-402. doi: 10.1104/pp.114.248963
COLLABORATIVE PROJECTS
## La isla de tomate
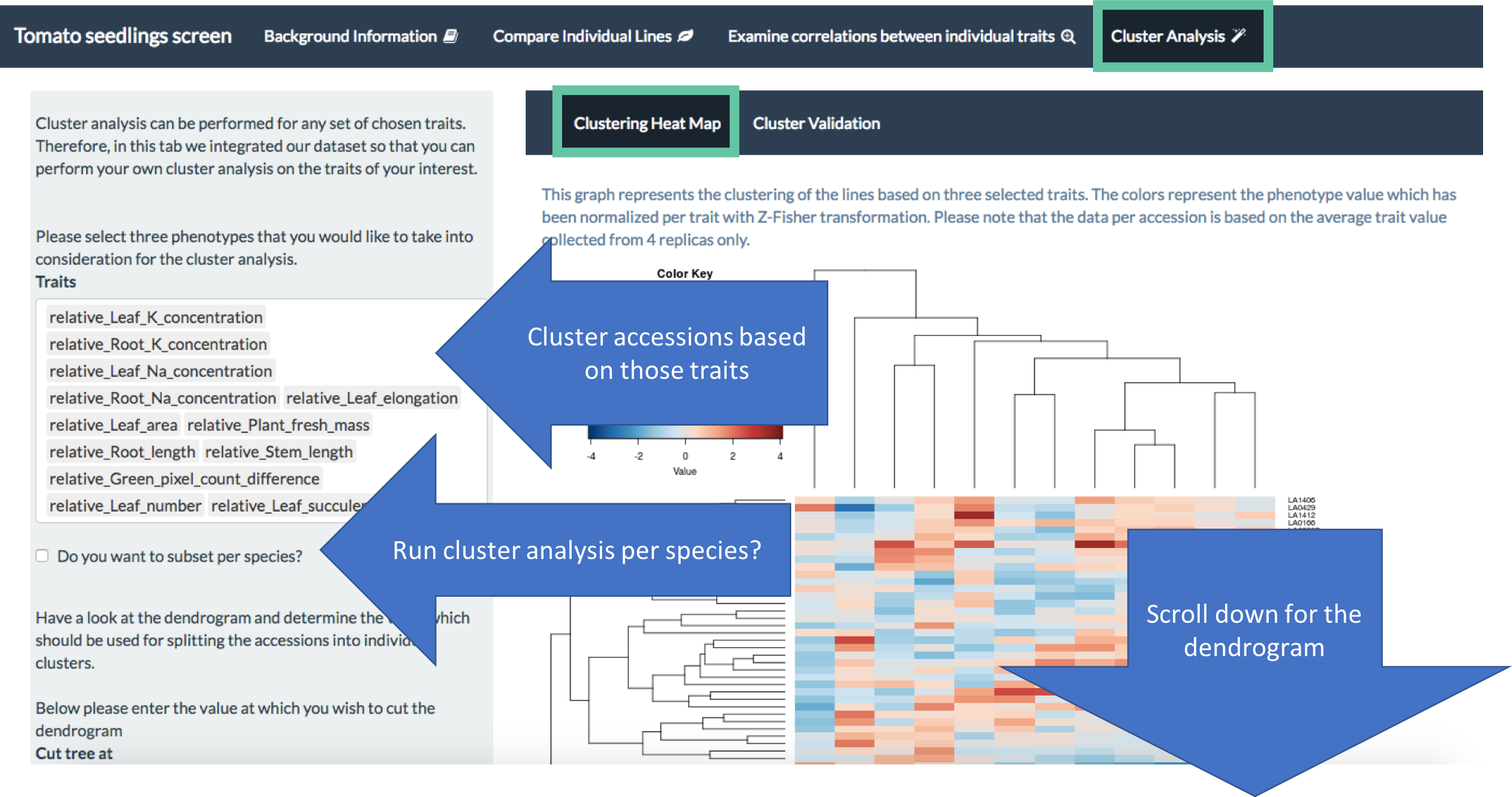
Ever wondered how salt tolerant the tomatos from Galapagos Island are? A PhD student in KAUST, Yveline Pailles, explored the salt stress responses of Galapagos accessions of two tomato species Solanum cheesemanea and Solanum galapagense. I helped her with some data analysis and designed an interactive data exploration tool that is available here. Instructions on how to use the app are available here, and the code can be found here. DOI: 10.5281/zenodo.1226455
## AI roots

Quantification of the Root System Architecture is not an easy task. Especially when you want to quantify the roots throughout time. We have a gigantic dataset consisting of over 4000 images of Arabidopsis thaliana roots including quantification of the most informative parameters, like Main Root Length, Lateral Root Density, Total Root Size and Shoot Length. Together with Andrew Yip, we are setting up a platform for deep learning for automatically quantify Root System Architecture from the images. The video can be found here, and you can follow our progress on twitter or our website.
PROTOCOLS:

For the protocols developed for the wet-lab as well as for in-silico data analysis, please see my protocols.io page.
LIST OF PUBLICATIONS:
- Julkowska, M.M., Mol, S., Klei, K., Hoefsloot, H., Feron, R., Tester, M., Keurentjes, J.B., Haring, M.A., de Boer, G., Testerink, C. (2017) Natural variation in Arabidopsis root plasticity reveals a role for HKT1-mediated ion transport in lateral root formation. Plant Cell doi: https://doi.org/10.1105/tpc.16.00680 - Article accompanied by the web-application.
- Awlia, M., Nigro, A., Schmoeckel, S., Negrao, S., Santielia, D., Trtilek, M., Tester, M., Julkowska, M.M., Panzarova, K. (2016) High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front. Plant Sci. Corresponding author
- Kawa, D., Julkowska, M.M., Montero-Sommerfeld, H., ter Horst ,A., Haring, M.A., Testerink, C. (2016) Phosphate-dependent root system architecture responses to salt stress. Plant Physiol. 172: 690-706.
- Thoen, M. P. M., Davila Olivas, N. H., Kloth, K. J., Coolen, S., Huang, P.-P., Aarts, M. G. M., Bac-Molenaar, J. A., Bakker, J., Bouwmeester, H. J., Broekgaarden, C., Bucher, J., Busscher-Lange, J., Cheng, X., Fradin, E. F., Jongsma, M. A., Julkowska, M. M., Keurentjes, J. J. B., Ligterink, W., Pieterse, C. M. J., Ruyter-Spira, C., Smant, G., Testerink, C., Usadel, B., van Loon, J. J. A., van Pelt, J. A., van Schaik, C. C., van Wees, S. C. M., Visser, R. G. F., Voorrips, R., Vosman, B., Vreugdenhil, D., Warmerdam, S., Wiegers, G. L., van Heerwaarden, J., Kruijer, W., van Eeuwijk, F. A. and Dicke, M. (2016) Genetic architecture of plant stress resistance: multi-trait genome-wide association mapping. New Phytol. 213: 1346-1362.
- Julkowska, M.M., Klei, K., Fokkens, L., Haring, M.A., Schranz, M.E., Testerink, C. (2016) Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J. Exp. Bot. 67(8): 2127-38.
- Julkowska, M.M., Testerink, C. (2015) Tuning plant signalling and growth to survive salt. Trends Plant Sci. 20: 586-594. Featured on Sept cover, accompanied by a video abstract
- Julkowska, M.M., Hoefsloot, H.C.J., Mol, S., Feron, R., de Boer, G-J, Haring, M.A., Testerink, C. (2014) Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol. 166: 1387-1402.
- Julkowska, M.M., McLoughlin, F., Galvan-Ampudia, C.S., Rankenberg, J.M., Kawa, D., Klimecka, M., Haring, M.A., Munnik, T., Kooijman, E.E., Testerink, C., (2014). Identification and functional characterization of the Arabidopsis Snf1-related protein kinase SnRK2.4 phosphatidic acid-binding domain. Plant Cell & Environ. 38(3): 614-24
- Wei, Z., Julkowska, M.M., Laloe, J-O, Hartman, Y., de Boer, G-J., Michelmore, R.W., van Tienderen, P.H., Testerink, C., Schranz, M.E. (2014) A mixed-model QTL analysis for salt tolerance in seedlings of crop-wild hybrids of lettuce. Mol. Breeding 34(3): 1389-1400
- Julkowska, M.M., Rankenberg, J.M., Testerink, C. (2013). Liposome-binding assays to assess specificity and affinity of phospholipid-protein interactions. Methods Mol. Biol. 1009: 261-271.
- Galvan-Ampudia, C. S., Julkowska, M. M., Darwish, E., Gandullo, J., Korver, R. A., Brunoud, G., Haring, M.A., Muunik, T., Vernoux, T., Testerink, C., (2013). Halotropism is a response of plant roots to avoid a saline environment. Current Biol. 23: 2044–2050.
- Jacobsen, J. V., Barrero, J. M., Hughes, T., Julkowska, M., Taylor, J. M., Xu, Q., & Gubler, F. (2013). Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta, 238: 121–138.
- McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., Lauriere, C., Munnik, T., Haring, M.A., Testerink, C., (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72: 436–449.
- Pribat, A., Sormani, R., Rousseau Gueutin, M., Julkowska, M. M., Testerink, C., Joubès, J., Castroviejo, M., Laguerre, M., Meyer, C., Germain, V., Rothan, C., (2012). A novel class of PTEN protein in displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem. J. 441: 161–171.